BLD Insights
Application of Linkers in Chemical Biology
12.11.2021
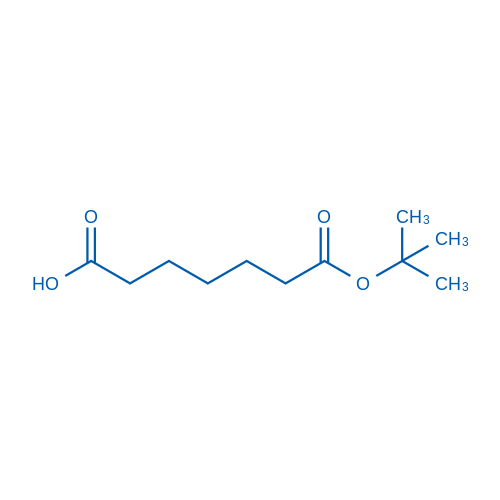
7-(tert-Butoxy)-7-oxoheptanoic acid
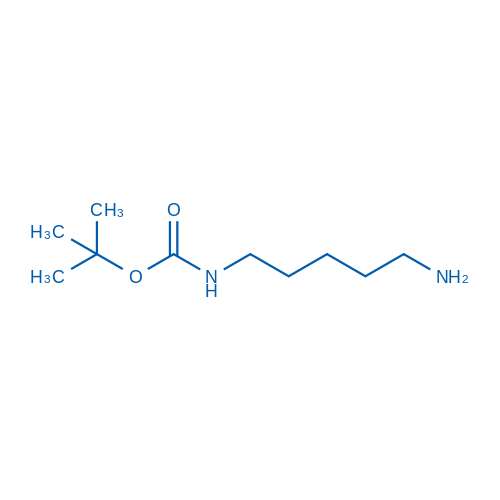
tert-Butyl (5-aminopentyl)carbamate
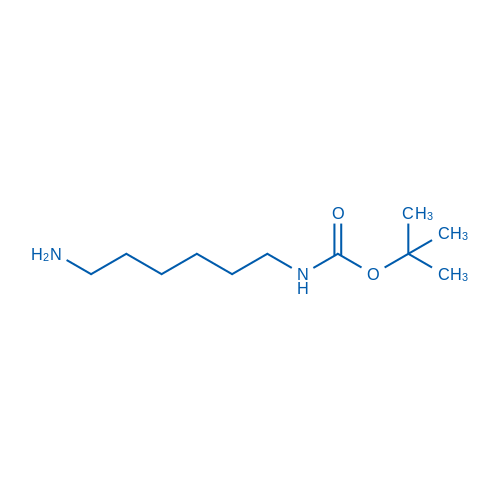
N-Boc-1,6-Diaminohexane
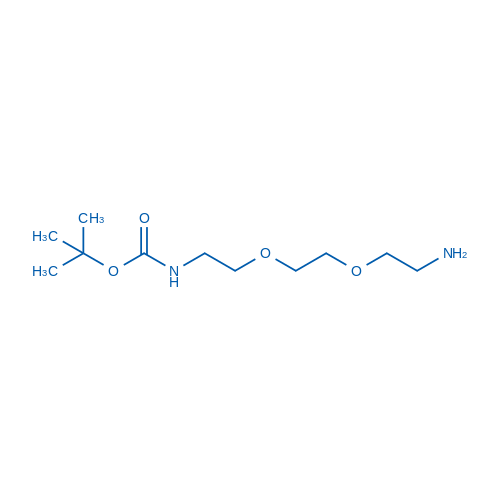
tert-Butyl (2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamate
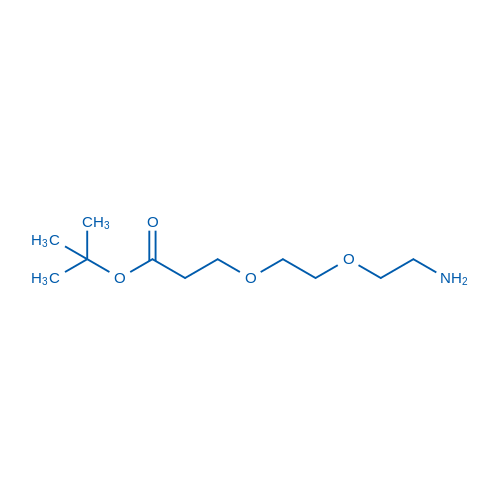
tert-Butyl 9-Amino-4,7-dioxanonanoate
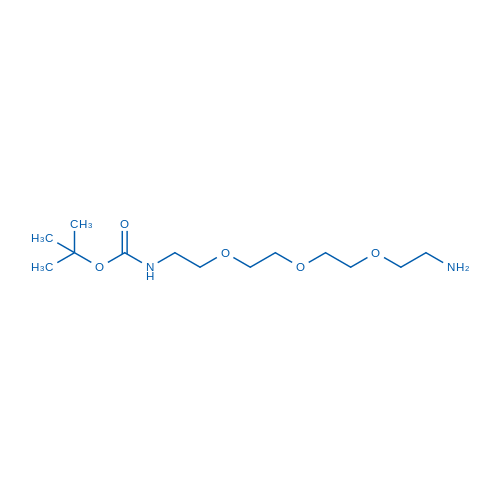
tert-Butyl (2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)carbamate
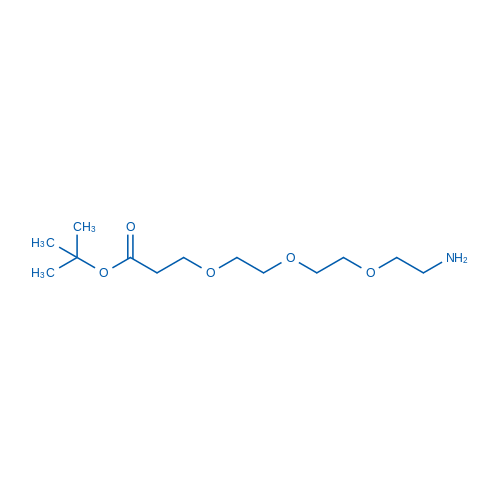
tert-Butyl 3-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)propanoate
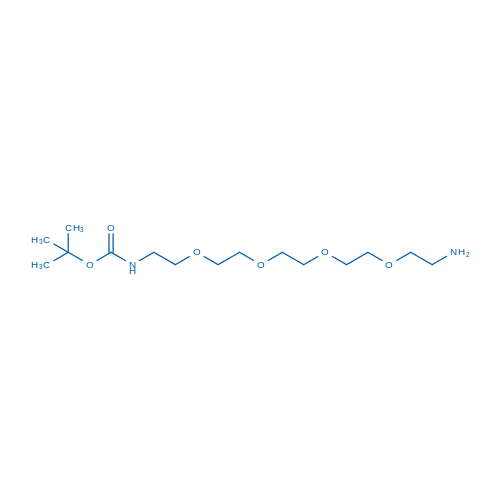
tert-Butyl (14-amino-3,6,9,12-tetraoxatetradecyl)carbamate
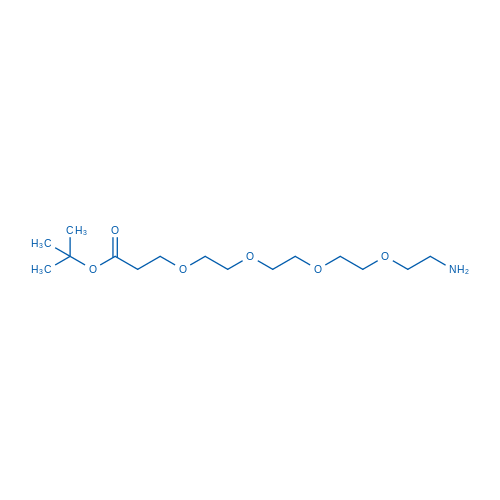
tert-Butyl 1-amino-3,6,9,12-tetraoxapentadecan-15-oate
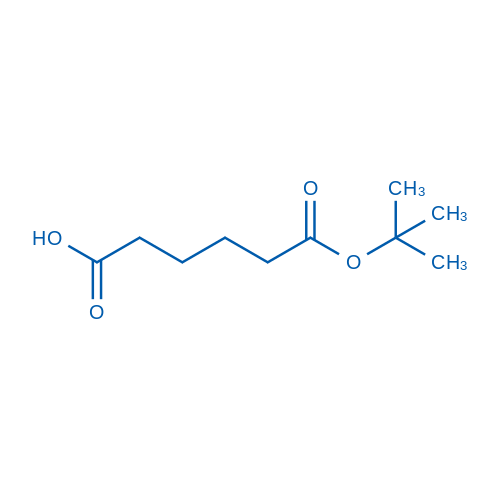
6-(tert-Butoxy)-6-oxohexanoic acid

7-(tert-Butoxy)-7-oxoheptanoic acid

tert-Butyl (5-aminopentyl)carbamate

N-Boc-1,6-Diaminohexane

tert-Butyl (2-(2-(2-aminoethoxy)ethoxy)ethyl)carbamate

tert-Butyl 9-Amino-4,7-dioxanonanoate

tert-Butyl (2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)carbamate
Linkers are often used as "bridges" for bifunctional molecules, and they play an important role in such molecules' structural and functional design. At the same time, choosing the right linker also can improve the effects in the optimization strategy of small molecule inhibitors.
Linkers are most widely used in the design and synthesis of PROTACs. PROTACs (Proteolytic Targeting Chimera) are hybrid bifunctional small molecules that consist of a warhead which binds to a target protein (POI) and a ligand that binds to E3 ligase, both of them are connected by flexible linkers. The choice of a right linker is of great significance in the process of PROTACs' optimization, it can not only regulate the appropriate spatial distance between the target protein and E3 ligase, but also determine the membrane permeability of the PROTAC molecules and the formation of the optimal conformation of the ternary complex. The popular cancer-causing mutant BRAF (V600E) drug has shown effective clinical effects, but the drug resistance caused by long-term administration has severely hindered the treatment of related diseases, in this regard, the Sicheri group of the University of Toronto designed and synthesized a series of PROTACs to inhibit the kinase activity of mutant BRAF[1], they use different types and lengths of linkers to connect the E3 ligase ligand and BRAF small molecule inhibitor to obtain a series of effective PROTACs, the most effective one is P4B, which in BRAF (V600E) Cell lines shows superior specificity and drug resistance compared to non-PROTAC controls.
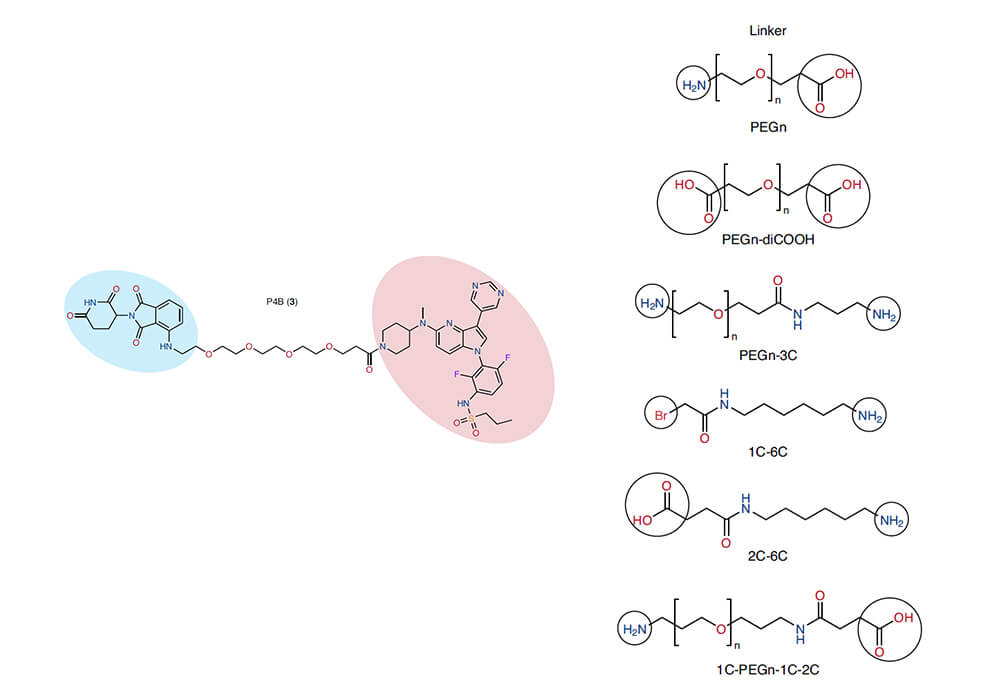
Figure 1. BRAF (V600E) PROTACs structural optimization
Besides, the addition of linkers can also solve the problems of drug resistance of conventional small molecule inhibitors. Li group at the State Key Laboratory of Biotherapy of Sichuan University has reported ConB-1[2], an inhibitor covalently binds to the 1259Cys outside the active site of ALK, which can effectively improve the drug resistance of ALK. The main work is to use different types of alkyl diethyl amine linkers to connect the covalent warhead and ceritinib. At the same time, the addition of linkers has significantly improved the molecule's anti-cancer activity and drug resistance, making the covalent inhibitor ConB-1 a promising clinical drug candidate for the treatment of NSCLC.
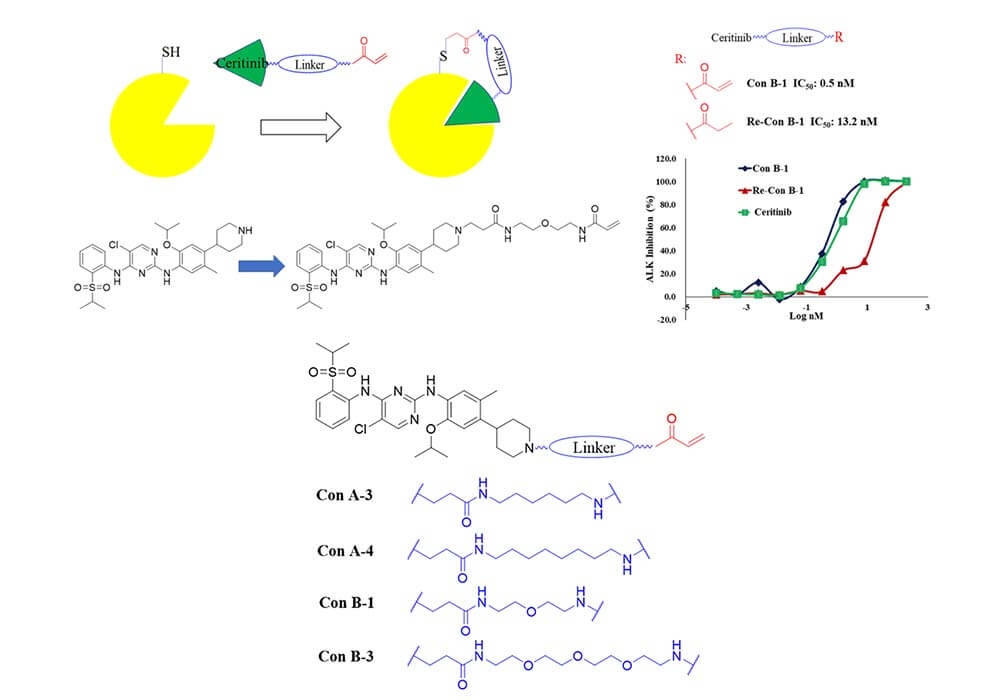
Figure 2. Binding mode and structural optimization of ALK covalent inhibitors
Linkers can also be used for the synthesis and development of PROTACs based on protein fusion tags. The Fischer group of Harvard Medical School reported a newly developed heterobifunctional degrader based on a "bump-and-hole" approach named XY-06-007, along with the corresponding protein degradation tag BRD4BD1L94V[3], the combination of them makes the degrader XY-06-007 more accurate and effective when targeting the substrate domain BRD4BD1L94V, which greatly reduces the off-target effect compared to the same type of other degraders. The key process of degrader XY-06-007's optimization is the selection of a suitable linker, in this work, the author tried several lengths of alkyl diethyl amine and PEG-derived linkers to connect the BET bromine domain inhibitor with the CRBN ligand to obtain a series of compounds, and finally got the most effective one: XY-06-007.
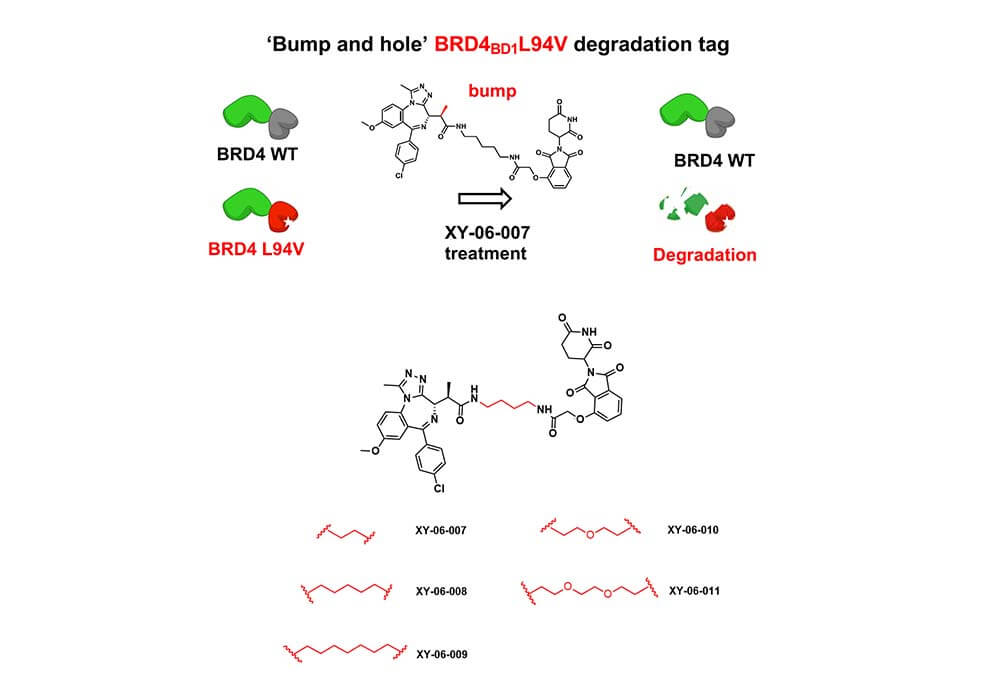
Figure 3. BRD4 degradation mechanism and structural optimization of degraders
References
[1]Posternak G, Tang X, Maisonneuve P, et al. Functional characterization of a PROTAC directed against BRAF mutant V600E [J]. Nat Chem Biol, 2020, 16(11): 1170-1178.
[2]Yan G, Zhong X, Pu C, et al. Targeting Cysteine Located Outside the Active Site: An Effective Strategy for Covalent ALKi Design [J]. J Med Chem, 2021, 64(3): 1558-1569.
[3]Nowak R P, Xiong Y, Kirmani N, et al. Structure-Guided Design of a "Bump-and-Hole" Bromodomain-Based Degradation Tag [J]. J Med Chem, 2021, 64(15): 11637-11650.
Prev: BLD Insights | MIDA Boronate: A New Organo-boron Reagent
Next: BLD Insights | The Utilization of Spirocyclic Scaffolds in Medicinal Chemistry